The Fondation Weizmann.be pour la science is a non-profit, private entity. Its aim is to promote the Weizmann Institute of Science in Belgium and to support its research projects via appropriate public relations activities as well as selective fund raising, in particular in the form of sponsorships, donations and legacies.
One of the Foundation’s regular activites is the sponsoring of promising students attending Belgian secondary schools, who qualify for participation in the annual 4 weeks International Scientific Summer School (ISSI) on the Weizmann Institute of Science campus, before continuing their tertiary education in science. This year two promising young women have been selected.
The Foundation is chaired by Mr Christian Hendboeg.
Mrs Diane Culer, Prof. Pierre Klees, Prof Maurice Sosnowski, Mr Paul de Schietere de Lophem, Mr Eric Hemeleers and Mr Roland Louis are directors.
The Belgian Foundation was founded in 2006, replacing the Belgian Committee, which had been established in 1973 by Prof. Georges Schnek, who also served as the first Secretary General until 1979. He was succeeded by Mr Louis Culer, who held this position for 20 years until 1999, followed by Prof. Marc van Montagu until June 2006.
Among the former Chairmen of the Committee were the former Belgian Prime Minister Theo Lefèvre (1973-1975), Prof. Piet de Somer, Rector of the “Katolieke Universiteit Leuven” (1975-1980) and Prof. Jean Brachet (1980-1972). Two Nobel Laureates, Prof. Christian de Duve and Prof. Ilya Prigogine, were members of the Academic Council.
Training Future Physician-Scientists: The Weizmann Institute Receives Approval for a New Medical School
by Christian Hendboeg | 16 February 2025 | Non classé | 0 Comments
The vision: Bridging the gap between today’s science and tomorrow’s medicine
Whether it’s artificial intelligence, computational biology or medical engineering, the worlds of medicine and science are facing a whole series of revolutions. In an attempt to address these emerging challenges, the Weizmann Institute of Science has developed a unique program to train the next generation of physician-scientists. This innovative study program has now been approved by Israel’s Council for Higher Education, and the Miriam and Aaron Gutwirth Medical School is expected to open its doors in October 2025.
By its very nature, the world of science is advancing at a faster pace than the world of medicine. The initiative to establish the new program was born out of a pressing need to bridge the gap between today’s science and tomorrow’s medicine. The program’s innovative approach will be to unify and intertwine its clinical and research components. The study groups will be small, and most of the time will be devoted to practice and research rather than to theoretical lectures. The multidisciplinary nature of Weizmann research will allow students to specialize not only in the life sciences but also in physics, chemistry, mathematics and computer science, while having access to the most advanced scientific infrastructure in the world. In parallel with conducting research at Weizmann, students will undergo clinical training at leading medical centers in Israel, including the Sheba Medical Center, Tel Aviv Sourasky Medical Center (Ichilov), Rambam Health Care Campus and Samson Assuta Ashdod Hospital, as well as hospitals run by the Clalit Group.

The program is expected to annually enroll up to 40 Israeli students possessing undergraduate or graduate degrees, who upon completion of their training will be eligible for a dual degree: a doctorate in medicine and a doctorate in research (MD-PhD). Registration is now open and, as in other study programs at the Weizmann Institute, no tuition fees will be charged and all students will be eligible for a scholarship.
“There is a growing need for physicians who have an in-depth understanding of basic and applied science, and have the tools and ability to translate scientific discoveries into new, improved medical treatments,” say Prof. Ayelet Erez and Prof. Liran Shlush – both heads of research groups at the Weizmann Institute and practicing physicians, who will lead the program on behalf of the Weizmann Institute. “The vision is to create a new generation of physician-scientists who are leaders in medicine and biomedical research. These will be professionals who are able to identify urgent clinical challenges, offer patients the opportunity to participate in clinical trials, and design and conduct innovative research that will lead to groundbreaking therapies – all with the aim of promoting public health and improving the quality of life of patients. Our physician-scientists will learn to delve deeper into problems and find creative solutions, while looking at medicine comprehensively – from identifying changes in large patient populations to the level of a single molecule in a specific patient.”
“From broad population trends to single molecules, our physician-scientists will tackle medical challenges with depth and creativity”
Prof. Erez and Prof. Shlush are joined by Prof. Iris Barshack and Prof. Asaf Vivante from Sheba Medical Center; Prof. Dina Ben-Yehuda from Hadassah Medical Center; and Prof. Amir Shlomai from Rabin Medical Center – Beilinson, part of the Clalit Group, who will serve as senior management team members.
The training of new physician-scientists is expected to provide a significant and unique boost to Israel’s public health system. Supplying Israeli hospitals with highly qualified personnel whose broad training includes both science and medicine will promote biomedical research and its application in Israel and bring the most cutting-edge, innovative treatments to patients.

“Educating and training the next generation of physician-scientists reflects the vision of the Weizmann Institute of Science to harness the power of science for humanity’s benefit,” says Weizmann Institute President Prof. Alon Chen. “The physician-scientist is expected to solve problems for patients and health care systems by challenging conventions and using scientific tools and research thinking. The medical studies program at the Weizmann Institute will significantly contribute to the precision medicine revolution in Israel. By leveraging our capabilities and resources, we will place the pioneers of future science at the service of the country’s health system, academia and biomedical industry.”
We Know What You Ate: Detailed Protein Maps Assess Intestinal Health
by Christian Hendboeg | 31 January 2025 | Non classé | 0 Comments
A new Weizmann Institute study identified all of the proteins in a stool sample – those from the microbiome, the human body and food – revealing the hidden secrets of the intestines and their impact on human disease
If the organs in our bodies could talk, the intestines might be the ones to divulge the most hidden truths about our lifestyle and health. Along the way, their “confessions” could supply crucial information for biomedical and clinical research. Weizmann Institute of Science researchers have now given just this sort of “voice” to the intestine. In a study being published today in Cell, the scientists present a method that can simultaneously identify, through testing a stool sample, all the proteins in the intestine – including those from food, from the person’s own body and from the intestinal microbiome. The method thereby makes it possible to decode the interactions among these proteins with unprecedented accuracy and resolution.

The microbiome was the starting point for the research, co-led by Drs. Rafael Valdés-Mas, Avner Leshem and Danping Zheng from Prof. Eran Elinav’s lab, in collaboration with Dr. Alon Savidor of the Nancy and Stephen Grand Israel National Center for Personalized Medicine. “We wanted to go beyond DNA sequencing, the usual method of studying the microbiome,” says Elinav, of Weizmann’s Systems Immunology Department. “DNA can tell us which bacteria are present in the gut and point to their potential activity. Bacterial proteins, in contrast, can directly reveal whether these bacteria are active, which activity they perform and how their function affects the human body in health and disease.”
Identifying proteins is challenging because of their sheer multitude, compounded by similarities among proteins from different species. The 20,000 or so protein-making genes in the human genome, for example, give rise to millions of protein variations; identifying them using existing protein databases can be complicated and highly time-consuming. The new Weizmann Institute method addresses this challenge, in part by combining DNA sequencing and mass spectrometry to generate a smaller, patient-tailored protein map.
“The proteins in the intestines are the ‘words’ that will one day reveal exactly what our intestines need – and how to provide it”
The method, dubbed IPHOMED – short for Integrated Proteo-genomics of HOst, MicrobiomE and Diet – makes it possible to decode the entirety of microbiome activity by showing which proteins in a stool sample come from which bacterial strains and in what amounts. In addition, it identifies the proteins secreted by the human gut in response to signals coming from the microbiome. Taken together, proteins from these two sources generate an atlas of the body’s communication with the microbiome, for example, during exposure to disease-causing bacteria or to antibiotics.
Thus, using the method, Weizmann researchers found that the human gut can secrete dozens of previously unknown antimicrobial peptides that act like natural antibiotics, killing some of the bacteria in the microbiome and ultimately shaping its makeup. This finding might help explain why the composition of each person’s microbiome is unique, leading to differences in susceptibility to disease.
No more cheating on the diet
When the researchers thought they’d finished developing the method, it could identify 97 percent of the proteins in each stool sample, which was a high rate, but the failure to consistently characterize the remaining 3 percent seemed puzzling. Further research clarified that these originated neither in the microbiome nor in the body tissues: They came from food.
This revelation suggested that the Weizmann method might be able to meet a dire, long-standing need of nutritional science, that of supplying a noninvasive means of revealing the exact details of a person’s diet. To address this challenge, the team created a database of proteins found in hundreds of food products and identified the ones unique to each food item. These developments made it possible to learn with unprecedented precision, from stool samples, what people had eaten. For example, when applied to samples collected from two groups of healthy volunteers, one in Germany and the other in Israel, the method identified a similar level of wheat consumption in both populations, but only the German samples had large amounts of proteins from pork; in contrast, most meat proteins in the Israeli samples came from poultry.
In one of the experiments conducted by the team, volunteers were asked to consume a changing repertoire of food items, including peanuts, on designated days. Not only did the method accurately determine when these food items were eaten, it was so sensitive that it could point to consumption of as few as five peanuts per day.
In another experiment, the researchers were able to accurately track changes in the diet of people with gastrointestinal diseases. In one case, the method correctly identified a child with newly diagnosed celiac disease who had failed to follow his prescribed gluten-free diet.
“Our method could be used to tell whether someone keeps kosher or whether a person is as strictly vegetarian as they proclaim themselves to be,” Elinav says with a smile. “But on a more serious note, the traditional diet-tracking method, self-reporting, is notoriously inexact. Knowing in greater precision and detail what people eat, even when their meal is complex and made up of multiple ingredients, can help establish which of the many components of a meal are beneficial to health and which are problematic.”
Advancing diagnosis and treatment of disease
To explore the use of their new method in the diagnosis and treatment of disease, the scientists applied it to stool samples of patients with inflammatory bowel disease, which is characterized by severe intestinal inflammation that is affected by diet and the microbiome. The analysis of samples from Israeli, German and American study participants enabled decoding, in great molecular detail, the altered interactions between the human gut and intestinal microbiome that drive the origins of this disease. The study led to the discovery of dozens of new proteins that could serve as potential future targets for drugs to treat this currently cureless disease. The researchers also identified human and bacterial proteins that, used together, could be developed into new biomarkers for diagnosing the type of the disease, assessing its severity and tracking its progress. These new probes promise to outperform calprotectin, the only clinically approved biomarker for inflammatory bowel disease.
In addition, using an IPHOMED analysis of the patients’ diet, the researchers were able to quantify the patients’ compliance with nutritional therapies for intestinal bowel disease and link the level of their adherence to such diets to improved control of inflammation. Moreover, they managed to apply their noninvasive method to detecting disease in the small intestine, the long, thin tube that in healthy people absorbs most of the proteins from food. Because the small intestine is notoriously difficult to visualize and access, this disease could not have been picked up by conventional means.
“Taken together, the proteins in the intestines are the ‘words’ that will one day allow us to hear exactly what our intestines are telling us and thus learn to give them exactly the help they need,” Elinav says. “This ability will help researchers devise personalized nutritional and medical interventions for a wide variety of disorders, particularly those affected by the microbiome, including inflammatory, metabolic, malignant and neurodegenerative diseases.”
Using the most recent high-throughput methods, IPHOMED simultaneously identified up to 15,268 bacterial proteins from the microbiome, up to 528 proteins secreted by the human body and up to 1,041 proteins from food.
Dr. Leshem, who was an MD-PhD student in Elinav’s lab during this study, is now a surgical resident at the Tel Aviv Sourasky Medical Center (Ichilov). Study participants also included Yotam Cohen, Dr. Lara Kern, Dr. Niv Zmora, Dr. Yiming He, Shimrit Eliyahu Miller, Tal Yosef Hevroni, Liron Richman, Barbara Raykhel, Shira Allswang, Reut Better, Fernando Slamovitz, Dr. Dragos Ciocan, Uria Mor, Dr. Mally Dori-Bachash, Shahar Molina, Dr. Jotham Suez, Dr. Suhaib K. Abdeen and Dr. Hagit Shapiro of Elinav’s lab; Dr. Merav Shmueli and Prof. Yifat Merbl of Weizmann’s Systems Immunology Department; Dr. Yishai Levin and Corine Katina of the Nancy and Stephen Grand Israel National Center for Personalized Medicine at the Weizmann Institute; Dr. Ghil Jona of Weizmann’s Life Sciences Core Facilities Department; Prof. Alon Harmelin and Dr. Noa Stettner of Weizmann’s Veterinary Resources Department; Dr. Aurelia Saftien, Dr. Nyssa Cullin, Kyanna S. Ouyang and Dr. Jens Puschhof of the Microbiome and Cancer Division, German Cancer Research Center (DKFZ), Germany; Prof. Koji Atarashi and Prof. Kenya Honda of the RIKEN Center for Integrative Medical Sciences (IMS) and Keio University, Japan; Prof. Nitsan Maharshak, Prof. Oren Shibolet and Prof. Zamir Halpern of the Tel Aviv Sourasky Medical Center (Ichilov), Israel; Prof. Dror S. Shouval and Prof. Raanan Shamir of Schneider Children’s Medical Center, Israel; Dr. Michal Kori of the Kaplan Medical Center, Israel; Prof. Minhu Chen of the First Affiliated Hospital, Sun Yat-Sen University, China; and Dr. Wolfgang Lieb, Dr. Corinna Bang and Prof. Andre Franke of the University Hospital of Schleswig-Holstein, Christian-Albrechts-University, Germany.
Couple therapy
by Christian Hendboeg | 5 October 2024 | Non classé | 0 Comments
A new Weizmann Institute method provides an unprecedented glimpse into the relationship between bacteria and their host cells
Like people, bacteria have their preferences when it comes to relationships. Some are totally independent, while others prefer company. Salmonella and many other kinds of bacteria are of the social type: They can live and even thrive inside a host cell. But unlike us, these bacteria do not spend a long time wooing the cell in the hope that it will welcome them in. Instead, they inject proteins that take control of the host cell’s systems.

In recent years, thanks in part to studies conducted by Prof. Roi Avraham’s team at the Weizmann Institute of Science, researchers have identified differences among the proteins that various bacterial subspecies inject into their hosts, which could explain why some of these subspecies are more virulent than others. For example, there are more than 2,500 subspecies of salmonella, but only a handful of them cause life-threatening disease. In a paper published in the Proceedings of the National Academy of Sciences (PNAS), scientists from Avraham’s team presented a new research method that shines fresh light on the relationship between bacteria and their host cells – and reveals what makes some bacteria particularly virulent.
Over the past decade, scientists have gained the ability to examine molecular processes at an unprecedented resolution, revolutionizing research in the life sciences. New methods of sequencing genetic material on the single-cell level have contributed to this revolution. But existing tools cannot be used to apply single-cell RNA sequencing to examine all the molecular relationships between thousands of bacterial subspecies and the no-less impressive array of hosts. To map the differences in virulence and disease-causing ability between the various subspecies of salmonella, for example, researchers need to sequence the DNA of the bacterial cells on the single-cell level, do the same for the DNA of the specific host cells that were infected and match up the findings of guest and host, which amounts to a daunting task.
“We discovered how bacteria manipulate host cells and how cells respond, which may help treat antibiotic-resistant infections”
In their study, the researchers applied their new method to 25 mutant salmonella species that infected immune system cells called macrophages. The method allowed them to study the relationship between each species of bacteria and its host cell and to identify a species that causes an exceptionally powerful immune response in the host. The species in question lacks a protein that the other bacteria express and successfully inject into the host. The researchers inferred that this protein is essential for repressing the host’s immune system. “We discovered a new role of a familiar protein and showed that it sabotages the host’s defense mechanisms,” Avraham explains. “In fact, bacteria inject many proteins into their hosts, and we still have not figured out which roles most of them play. Our method, which is already used by researchers around the world, will make it possible to continue systematically revealing these roles. Moreover, it can be applied to any kind of bacteria, including friendly bacteria that are vital to many of our body’s systems.”
Beyond advancing basic research, the new method might help develop ways of battling bacterial resistance to antibiotics, defined by the World Health Organization as one of the main threats to human health and food security. “There are two new potential defense strategies, both still in the early stages of development,” Avraham says. “One is reducing the virulence of disease-causing bacteria and the other is bolstering the immune response of the host cells. Our method makes it possible to study both: simultaneously understanding how the bacterium launches its attack and how the host cell defends itself.”

It is estimated that by 2050, around 10,000,000 people a year will die as a result of ever-increasing bacterial resistance to antibiotics – close to the number of people who died of cancer worldwide in 2020.
Also participating in the study were Dror Yehezkel, Dr. Camilla Ciolli Mattioli, Neta Blumberger, Dr. Gili Rosenberg, Aryeh Solomon and Dr. Dotan Hoffman from the Immunology and Regenerative Biology Department at the Weizmann Institute.
Peeling Back the Layers of Brain Tumors
by Christian Hendboeg | 30 August 2024 | Non classé | 0 Comments
Weizmann scientists have mapped common types of brain tumors at unprecedented resolution – and identified a possible reason why some patients fail to respond to a new drug
The cells that make up cancerous brain tumors are extremely varied and sometimes create unique three-dimensional shapes. As far back as 1932, American neurosurgeon Percival Bailey attempted to label these cells and discovered that they can be divided into several families of cells with similar properties. More than ninety years later, we still know precious little about the identities of cell groups that make up different kinds of brain tumors, these groups’ organization and how they affect the course of the disease and the outcome of treatment. This is why the success rate for treatment of most brain cancers is typically not high.
Over the past decade, genetic sequencing technology that works at the single-cell level has made it possible to examine, in minute detail and in one fell swoop, thousands of cells in the same tissue, to understand which genes they express and to then categorize them and study the role of each group. Scientists in Dr. Itay Tirosh’s research group in the Weizmann Institute of Science’s Molecular Cell Biology Department, in collaboration with Prof. Mario L. Suvà’s lab at Massachusetts General Hospital, harnessed this technology in order to reexamine some of the unanswered questions in the field of brain tumors.

The most common type of primary brain tumor is the glioma, which originates from the support cells that assist our nerve cells. There are two main types of glioma tumors: those that are usually less aggressive and have a mutation in the gene encoding an enzyme called IDH, and those without this mutation, which are highly aggressive and known in medical terminology as glioblastoma. In the past few years, researchers from Tirosh’s lab have been using single-cell RNA sequencing to analyze the cellular composition of both kinds of tumors. They revealed that the tumor cells are divided into groups, each of which expresses a unique genetic program that determines the biological “state” of the cancer cells in this group. Among other findings, the researchers discovered groups of cells that use their unique genetic programs to mimic normal brain cells.
In a new study published recently in Cell, researchers from Tirosh’s lab – led by Dr. Alissa Greenwald, Noam Galili Darnell and Dr. Rouven Hoefflin – harnessed technologies that make it possible to not only sequence the RNA on the single-cell level but also spatially map its expression. This allowed them, for the first time, to identify which genes are uniquely expressed in each of the thousands of areas within a brain tumor. As a result, they were able to precisely map how glioblastoma and glioma tumors are organized. To conduct the study, they took biopsies from 13 patients with glioblastomas and from six patients with gliomas that had the IDH mutation.
The driving force behind the tumor’s layered structure is the lack of oxygen, which is exacerbated as the disease progresses and the tumor develops
The researchers’ first discovery was that the groups of various cells within a glioma tumor are not distributed evenly across the tumor; rather, they are concentrated in various environments inside the growth. These microenvironments are not entirely homogenous: Cells from other groups were always found in proximity to other types of cells. In the next stage of the study, the researchers checked whether there were groups of tumor cells that usually exist in proximity to each other. They discovered that the cells not only had preferred neighbors but also that these good-neighbor couplings were consistent in different patients.
Certain neighboring pairs imitated the natural behavior of brain tissue. For example, cells that imitate the parent cells of the oligodendrocyte support cell were found close to endothelial cells, which line the walls of blood vessels. This coupling also occurs in healthy tissue, since endothelial cells release substances that are vital for the survival and proliferation of oligodendrocyte precursor cells. Similarly, cells that imitate neuron progenitor cells were found in the parts of the tumor that penetrated healthy brain tissue, just as progenitor cells in healthy tissue migrate when the tissue is regenerated.
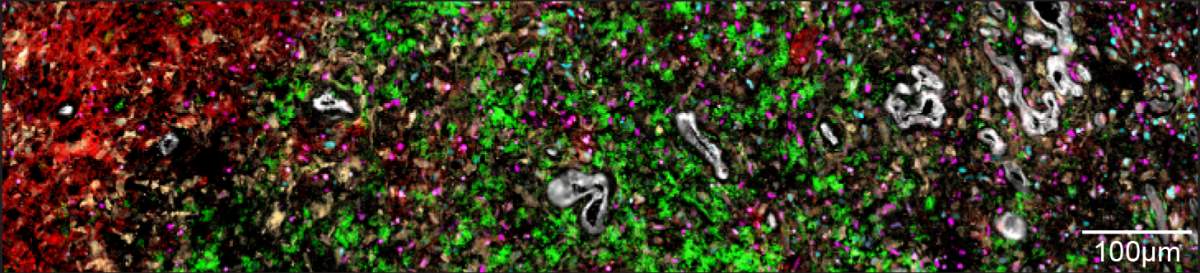
Taking an overview to gain a fuller understanding of these couplings, the researchers realized that the cells created five distinct layers by organizing themselves into separate environments within the tumor. The innermost layer – the core of the tumor – is made up of necrotic cells, which do not receive enough oxygen to survive. In the layer surrounding the necrotic core, the researchers found cells similar to embryonic connective tissue, as well as additional cells, including immune system cells responsible for causing inflammation. The third layer was primarily made up of blood vessels, endothelial cells forming blood vessel walls and additional immune system cells.
Cells in the two outer layers of the tumor don’t suffer from a lack of oxygen. This enables groups of tumor cells that mimic healthy brain tissue – progenitors of neurons and support cells – to develop in the fourth layer. The fifth, outermost layer contains healthy brain tissue, into which the tumor penetrates. These findings about the different layers of a tumor indicate that the driving force behind the tumor’s layered structure is the lack of oxygen, which is exacerbated as the disease progresses and the tumor develops.
Based on these findings, the researchers noticed a much more chaotic structure in less aggressive tumors – which are also usually smaller – and in areas of the tumor with a plentiful supply of oxygen. In most glioma tumors with the IDH mutation, for example, there was usually no necrotic tissue, and the structure of the tumor was disorganized; in the rare cases when there was necrotic tissue, the biopsies also showed a relatively well-ordered structure.
“We discovered that an organized spatial structure is characteristic of the more aggressive tumors,” Tirosh explains. “The lack of oxygen in the tumor cells’ environment influences the gene program that they express and therefore affects their state. As the tumor grows, distinct layers are formed, some of which may be less accessible to drugs and to cells from the immune system, and these could make the tumor more resilient.”
The changing status of cancer cells
Researchers from Tirosh’s lab used the information they collected on the cellular composition of glioma tumors to work out how a new, promising drug helped some of the patients with this type of cancer. To do so, they used biopsies from tumors of three patients who had participated in a clinical trial of the new drug and who had responded to the treatment, as well as biopsies from six patients who had not undergone any treatment. To complete the picture, they also used data from biopsies taken from an additional 23 patients who had taken the drug and 134 patients who had not.
The research team, led by Dr. Avishay Spitzer, found that the drug, which works by inhibiting the mutant IDH enzyme, caused the cells to alter the gene program that they expressed. In fact the treatment encourages the cancerous stem cells to differentiate into mature cells, thereby undermining their ability to divide rapidly, blocking the disease’s progress.

The researchers postulated that if the drug works by causing cancerous cells to differentiate into mature cells, the mutation attacking the gene that is critical to the differentiation process could explain those cases in which the drug does not work. In the biopsies taken from patients who did not receive the drug, they identified a certain gene that is linked to low levels of mature cancerous cells. When they silenced that gene in a mouse model of cancer, they found, as expected, that the drug did not work. “This indicates that the gene mutation we identified could be a biological marker allowing us to determine in advance which patients will benefit from the treatment and which will not,” Tirosh explains. These new findings could also help find a course of treatment that combines IDH inhibitors with another drug that encourages the differentiation process and increases the treatment’s impact on the tumor.
“Our two most recent studies revealed the forces that shape the character of cancerous cells in a tumor, be that in their untouched environment or in one resulting from a therapy that alters the cells’ genetic program,” Tirosh says. “These findings pave the way for a new approach to cancer treatment, since once we are familiar with the cell groups that populate every area of the tumor and we know how a cell can move from one state to another, we might be able to develop new targeted treatments that will alter the course of the disease. The understanding that both the composition of cells within the tumor and its three-dimensional structure are linked to the level of the tumor’s aggressiveness could also lead to new diagnostic methods that do not rely solely on the volume of the tumor and the mutations it contains.”
The median survival time for people diagnosed with glioblastoma is just 14 months; for those diagnosed with grade 2 oligodendroglioma – the mutant IDH glioma – it is 11 years.
Participants in the structural study of glioma also included Dr. Dor Simkin, Yotam Harnik and Dr. Julie Laffy from Weizmann’s Molecular Cell Biology Department; Dr. Christopher W. Mount, Dr. Nicolas Gonzalez Castro, Sydney Dumont, Dr. Masashi Nomura and Vamsi Mangena from the Massachusetts General Hospital and Harvard Medical School in Boston, MA, USA; Dana Hirsch from Weizmann’s Veterinary Resources Department; Tom Talpir from Weizmann’s Computer Science and Applied Mathematics Department; Dr. Merav Kedmi, Dr. Inna Goliand, Dr. Hadas Keren-Shaul and Dr. Yoseph Addadi from Weizmann’s Life Sciences Core Facilities Department; Gioele Medici, Prof. Michael Weller and Dr. Marian C. Neidert from University Hospital Zurich, Switzerland; and Dr. Baoguo Li from Weizmann’s Systems Immunology Department.
Participants in the IDH inhibitor study also included Dr. Rony Chanoch-Myers from Weizmann’s Molecular Cell Biology Department; Dr. Simon Gritsch, Dr. Masashi Nomura, Hannah R. Weisman, Dr. Nicolas Gonzalez Castro, Nicholas Druck, John J. Y. Lee, Ravindra Mylvaganam, Rachel Lee Servis, Jeremy Man Fung, Prof. Christine K. Lee, Dr. Hiroaki Nagashima, Prof. Julie Miller, Dr. Isabel Arrillaga-Romany, Dr. David N. Louis and Prof. Hiroaki Wakimoto from the Massachusetts General Hospital and Harvard Medical School in Boston, MA, USA; Dr. Jerome Fortin from Princess Margaret Cancer Centre in Toronto and McGill University in Montreal, Canada; Dr. Ramya Raviram, Prof. Dan A. Landau and Dr. Daniel P. Cahill from the New York Genome Center; Will Pisano and Prof. Keith L. Ligon from Brigham and Women’s Hospital in Boston, MA, USA; Prof. Patrick Y. Wen from the Dana-Farber Cancer Institute, Harvard Medical School, Boston; Prof. Tak W. Mak from the University of Hong Kong; and Prof. Marc Sanson and Dr. Mehdi Touat from the Sorbonne University, Paris.
Dr. Itay Tirosh’s research is supported by the Moross Integrated Cancer Center, the Zuckerman STEM Leadership Program and the Rising Tide Foundation. Dr. Tirosh is the incumbent of the Dr. Celia Zwillenberg-Fridman and Dr. Lutz Zwillenberg Career Development Chair.
The Weizmann Institute of Science Ranked among the World’s Top Ten Academic Institutions
by Christian Hendboeg | 30 August 2024 | Non classé | 0 Comments
The Weizmann Institute of Science maintains its status as one of the world’s leading research institutions. It has recently been placed in the top ten among universities worldwide in a weighted (proportional) ranking of research quality published annually by the Centre for Science and Technology Studies (CWTS) of Leiden University, the Netherlands (“Leiden Ranking”). This year Weizmann is ranked tenth for general research quality – alongside institutions such as Princeton, Harvard, Stanford and MIT – and sixth in the world for research quality in the biomedical and health sciences.

The Leiden Ranking, published since 2007, is purely quantitative and does not use subjective surveys. The data encompass the number of scientific publications by scientists from the various universities and research institutes, the number of citations for these publications – indicating the quality of the research – and additional information that takes into account the size of the institutions.
Additionally, the Weizmann Institute was recently placed among the top 100 in the 2024 Academic Ranking of World Universities (ARWU), released annually by ShanghaiRanking Consultancy. Weizmann was listed 69th, the highest ranking among Israel’s academic institutions; in chemistry and biology, it was ranked 44th and 45th, respectively. Unlike the Leiden Ranking, which focuses on the quality of research, the ARWU, which takes into account various parameters, is regarded as affording a built-in advantage to larger institutions over smaller ones. Still, the Weizmann Institute improved its ARWU standing in recent years, moving up from 83rd in 2022 to its current ranking.
- Between 2019-2022 Weizmann Institute scientists published 2,450 scientific papers that were cited 38,760 times.
- Overall, more than 63% of the Weizmann Institute’s scientific papers were included in the top 50% of publications with the greatest influence in their field for those years; 18% were included in the top 10% of publications; and 2.5% were in the top 1% of publications.
- In the biomedical and health sciences, more than 20% of the Weizmann Institute’s scientific papers were ranked in the top 10% of the most influential scientific publications.




