Understanding how the bacteria gets around a common drug may help develop new ones to combat a growing threat
The World Health Organization (WHO) recently issued a new warning about strains of pathogenic bacteria that are now resistant to all known antibiotics. The warning had made headlines around the world, raising the specter of “super-bugs” that have been developing resistance to most of the antibiotics used in the clinic. Clearly, advanced antibiotic drugs are desperately needed. But to achieve this aim, we first need to understand how antibiotics work – the exact molecular mechanisms by which they act, as well as the mechanisms of acquiring antibiotic resistance. New research at the Weizmann Institute of Science has solved a puzzle about a resistance to one of the oldest antibiotics on the shelf, erythromycin.
Discovered in the 1920’s, antibiotics were first widely used at the end of the Second World War. They are not a human invention but rather a part of the natural defense mechanisms of bacteria: the weapons produced by bacteria for use against other species of bacteria in the wars of survival they wage against one another. Antibiotic resistance also exists in nature, as bacteria continue to evolve ways of evading these weapons. The problem is that the widespread use of antibiotics by humans has accelerated the appearance of bacteria that are resistant to these drugs, leading, today, to the incidence of “super-bugs” that are resistant to multiple antibiotics.

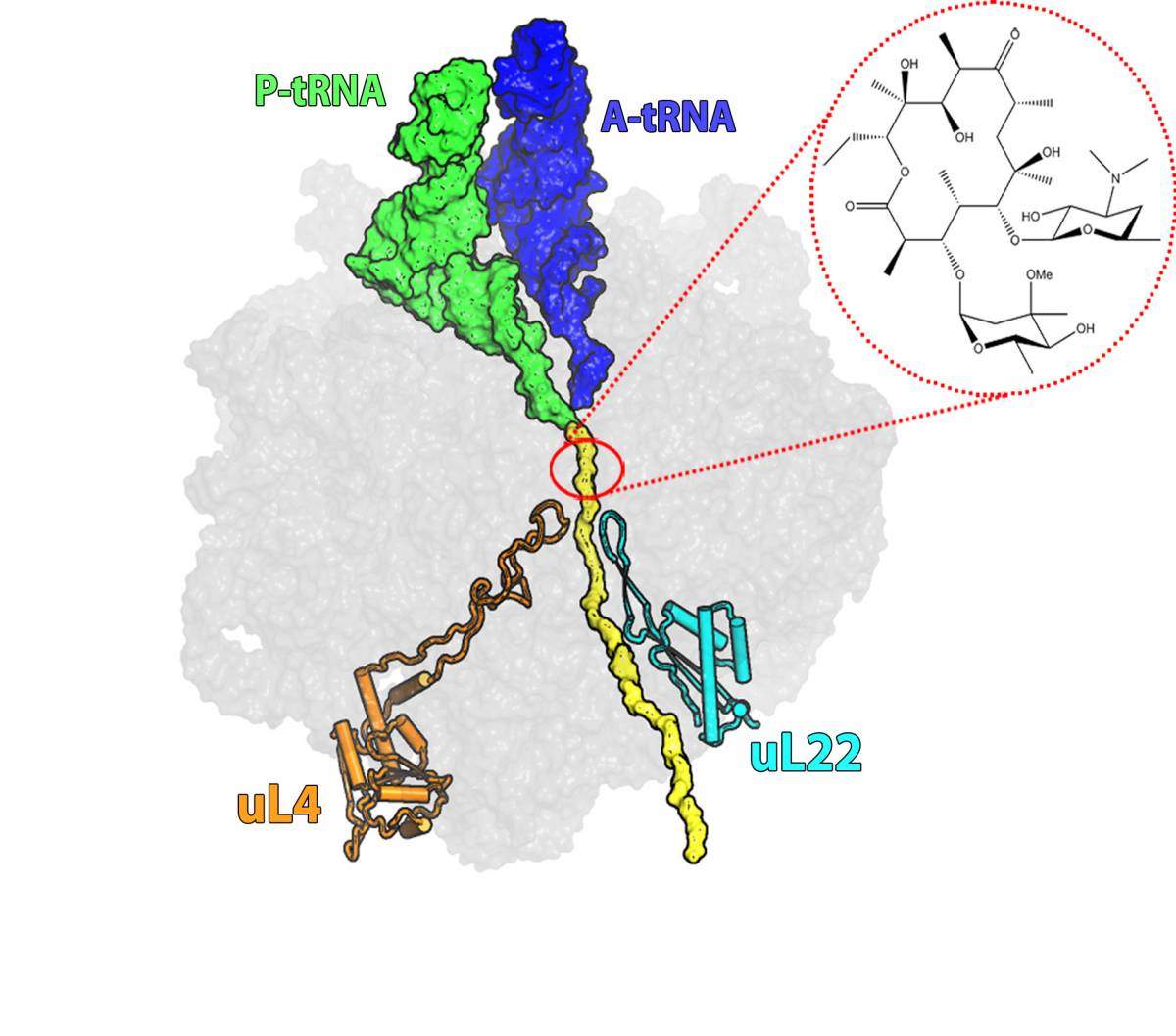
Erythromycin, first discovered in 1949 and still prescribed today, is one of these bacterial defense mechanisms. It belongs to the macrolide family of antibiotics, which works by binding to the ribosome – the cells’ protein factory. These antibiotics interfere with the ribosome’s function: without new proteins the cells cannot grow and multiply.
About 40% of antibiotics work through ribosome inhibition, but each has a different mechanism. Erythromycin, for example, binds to a site on the “protein exit tunnel” of the ribosome and blocks it, thus preventing the production of full length proteins or else hampering the release of new proteins from the ribosome into the cell. Most of the bacteria that have developed resistance to erythromycin have done so through a mutation that prevents the drug molecule from attaching itself to the drug’s binding pocket at the tunnel’s entrance. But some researchers had observed bacteria in which erythromycin was bound to its binding pocket, but the bacteria were, nonetheless, resistant to its effects. This resistance involved a different mutation — this one in a ribosomal protein called L22.
The mutated protein does not appear to interfere with the actions of the erythromycin; nevertheless bacteria with this mutation are resistant
Dr. Itai Wekselman, who solved this riddle as a part of his research in the lab of Nobel laureate Prof. Ada Yonath, says that the resistance mechanism was especially puzzling because, “the antibiotic molecule is not in contact with the mutated protein. By biochemical standards, the protein is located relatively far away from the antibiotic binding site. The mutated protein does not appear to interfere with the actions of the erythromycin; nevertheless bacteria with this mutation are resistant.”
In these studies, Wekselman, Dr. Anat Bashan, Dr. Ella Zimmerman, Dr. Haim Rozenberg and others in Prof. Yonath’s group in the Weizmann Institute’s Structural Biology Department, together with Dr. Janice Zengel of the University of Maryland and Prof. Hanne Ingmar of the University of Copenhagen, made use of three decades groundbreaking research in revealing the three-dimensional structure of the ribosome. They also benefited from advances in x-ray crystallography, which enabled them to envision these complex structures at near-atomic resolution – with and without the mutation, in the presence or the absence of erythromycin.

They discovered that the mutation increases the width of a loop at the tip of the L22 protein and moves it toward the center of the tunnel. This movement initiates a domino effect in which some of the RNA bases that make up the tunnel are relocated. These changes propagate through the tunnel – reaching up to the erythromycin binding pocket. Even though they do not prevent the antibiotic from binding, they still manage to keep it from blocking the tunnel by increasing the space available for growing polypeptides, allowing protein production to proceed.
More on mechanisms of resistance
In another study recently publish in mBio, led by Zohar Eyal, a PhD student, conducted in collaboration with other members of Yonath’s lab, together with Dr. Matthew Belousoff and Prof. Trevor Lithgow of Monash University, Australia, and Prof. Shashi Bhushan of Nanyang Technological University, Singapore, used new techniques in electron microscopy to investigate the molecular mechanisms of antibiotic resistance in Staphylococcus aureus, a bacterium responsible, among other things, for many hospital-acquired antibiotic-resistant infections. Dr. Anat Bashan says: “People are dying today of diseases like pneumonia – and not just in developing countries. These findings could enable the development of new antibiotic drugs that overcome these resistance mechanisms.”
Today, few new antibiotics are entering clinical use and, because the profit margins are slim and the costs of developing new drugs high, pharmaceutical companies are not investing sufficient resources in antibiotics R & D. Prof. Yonath has been sounding warnings on this issue for years: “If the drug companies do not act quickly, in another 20-30 years we could find ourselves back in the pre-antibiotic era.”
Prof. Ada Yonath’s research is supported by the Helen and Milton A. Kimmelman Center for Biomolecular Structure and Assembly, which she heads. Prof. Yonath is the incumbent of the Martin S. and Helen Kimmel Professorial Chair of Structural Biology.

Recent Comments