Self-assembling protein complexes based on a single mutation could provide scaffolding for nanostructures
The proteins in our bodies are social molecules. But now and again, new ties between proteins can get you into trouble. For example, when hemoglobin – the protein complex that carries oxygen in our blood – undergoes just one mutation, the complexes stick to one another, stacking like Lego blocks to form long, stiff filaments. These filaments, in turn, elongate the red blood cells found in sickle-cell disease. For over 50 years, this has been the only known textbook example in which a mutation causes these filaments to form. According to Dr. Emmanuel Levy and his group in the Weizmann Institute of Science’s Structural Biology Department, Lego-like assemblies should have formed relatively frequently during evolution, and so they asked: How easy is it to get proteins to stack into filaments? Their answer, which was recently published in Nature, may have implications for both biological research and nanoscience.
Levy and his group took their inspiration from the structural element that enables hemoglobin to stick in this way: symmetry. Symmetric protein complexes are made of identical units; if you flip the complex over, it will look the same, something like an Oreo cookie. And since identical units are produced from the same gene, each genetic mutation is repeated multiple times in the complex. If each complex is a “cookie” with sticky mutations on both sides, such complexes can continue to stick together, one after the other, ad infinitum. Sickle-cell filaments are unique in that the protein complexes within them retain their original shape. Other filaments, for example the amyloid filaments that form plaques in Alzheimer’s disease, require the proteins to change shape – partially unfolding – before they can join.
The stickiness occurs because the mutation substitutes an amino acid that is normally hydrophilic – “water-loving” – with one that is hydrophobic – “water-hating.” In the watery environment in which proteins move, the hydrophobic regions on those proteins prefer to interact with one another, like foam bubbles in water.
In their experiments, Levy and his group, including Hector Garcia-Seisdedos, Charly Empereur-Mot (who is now at Conservatoire National des Arts et Métiers in Paris) and Nadav Elad of the Weizmann Institute’s Chemical Research Support Department, began with an ultra-symmetric protein complex made up of eight identical units. “That way, for each mutation you introduce into the gene, you get an additional seven mutations for free,” says Levy. They followed just one rule for mutating the proteins: Switch a hydrophilic amino acid with a hydrophobic, “sticky,” one.
The team initially created proteins with three mutations to two different sticky amino acids and observed Lego-like self-assembly in both cases. Investigating further, the team experimented with each mutation individually and found that one was capable, on its own, of producing the long filaments. Electron microscopy confirmed that the filaments were, indeed, held together by the proteins’ sticky spots.
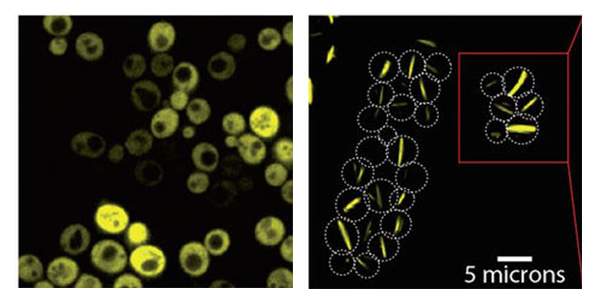
Following the successful design of this particular protein complex, the group approached their original question: Are mutations that only do one thing – increase the stickiness of the protein’s surface – likely to induce Lego-like self-assembly? They mutated 11 additional proteins that were known to be symmetric – creating 73 different mutations in all – and produced them in baker’s yeast cells, adding a fluorescent protein “label” to enable their visualization. In 30 of these variations, the researchers observed behavior that suggested self-assembly: Around half of these had stacked into long filaments, while the other half were bunched together in a more amorphous way, forming “foci.” The researchers then filmed the yeast cells as they grew under a microscope lens. The cells proceeded to produce neat bundles of filaments spanning entire cells, from end to end. “It’s amazing that the yeast cells, which have never seen this protein before, not only produce the fibers but manage to grow with them and snap them when two dividing cells separate,” says Levy.
Yeast cells producing a bacterial Lego-like protein complex
If the researchers reproduced the phenomenon of sickle-cell filaments so easily in the lab, why is it not seen more in biomedical research? There may be two answers to this question. Firstly, the team revealed that naturally symmetric proteins evolved to have extra hydrophilic amino acids on their surfaces, thus minimizing the risk of self-assembly. What was the basics of this experiment. Secondly, says Levy, researchers probably see more Lego assemblies than they think. The localized foci, in particular, may often be dismissed as “artifacts of misfolded proteins,” whereas in fact they are biologically relevant assemblies of folded proteins. “Now that researchers know they can evolve so readily, they may look at foci more carefully and see many more biologically relevant Lego-like assemblies.”
“Also,” he adds, “the filaments are produced so easily in the yeast, they could be good candidates for the scaffolding of nanostructures. Our study was unique in that it did not require complex computational design, nor did we have to scan thousands of mutations to find the one we wanted. We simply started with an existing structure and found a simple strategy to induce the assembly of filaments. We are continuing to research this phenomenon to understand exactly how it happens in nature as well as in artificially mutated proteins.”
Dr. Emmanuel Levy’s research is supported by the David and Fela Shapell Family Foundation INCPM Fund for Preclinical Studies; the Henry Chanoch Krenter Institute for Biomedical Imaging and Genomics; the Louis and Fannie Tolz Collaborative Research Project; the Richard Bar Laboratory; and Anne-Marie Boucher, Canada. Dr. Levy is the incumbent of the Recanati Career Development Chair of Cancer Research in Perpetuity.

Recent Comments