Consider the growing embryo: All its cells share the same genes, but they have entirely different fates, ultimately specializing to make up the brain, liver, kidneys or other body organs. It’s as if all the cells contain the same “book” – that is, the genome – but each cell “reads” only the chapters that dictate its identity. How do cells flip to the relevant “chapters”? As reported recently in Nature Biotechnology, Weizmann Institute scientists have developed a new method for deciphering “epigenetic tags”: coded instructions that determine cellular fate.

Dr. Ido Amit identifies two-word phrases in the genetic “instructions”
Epigenetic tags are issued by the molecular packaging and scaffolding of DNA, as well as by the genome itself. The tags – bits of chemical code – are attached to the protein spools around which the DNA is wound in the cell’s nucleus. The tags can “instruct” the DNA to unwind somewhat, enabling a portion of the genome to be read out, or, alternatively, cause the DNA to tighten around the spools, so that the genes in that section of the genome remain inactive. Epigenetic tags are much more susceptible to environmental pressures than the genome, and they thus continue to change throughout a person’s life. Mistakes in epigenetic markings, such as their improper placement, have been implicated in a variety of diseases: “Each cell in our body has a characteristic epigenetic identity,” says team leader Dr. Ido Amit of the Immunology Department. “The epigenetic machinery may contain numerous potentially cancer-causing alterations, and medications are currently being developed to target these epigenetic features.”
More than a hundred types of epigenetic tags have already been identified and profiled using a method known as ChIP-seq: Chromosomes are broken into fragments and treated – the technical term is “enriched” – with antibodies that label specific tags; the DNA is then sequenced to establish the tags’ location on the genome. This approach makes it possible to track one type of tag at a time. But scientists have been suggesting for a while that – something like compound words in our vocabulary – epigenetic tags sometimes work together to convey a new meaning. For example, when the “Read me!” and “Ignore me!” tags appear together, the resultant command is “Hold off!” Identifying such “bivalent” commands, however, poses a tremendous challenge, because millions of different tags are attached to a DNA molecule at any given time, the tags change over time and they vary from one cell to another, even within the same type of cells.
All its cells share the same genes, but they have entirely different fates
Now research students Assaf Weiner and David Lara-Astiaso, together with other members of Amit’s team, have developed a new technology, called co-ChIP, for revealing pairs of epigenetic tags that work together. “Simply performing two sequential steps, using two antibodies, required a huge amount of biological material and produced messy results that were difficult to interpret,” says Weiner. “So instead of treating a chromosome fragment with the first tag, we developed a technique for labeling it with a unique genetic barcode sequence. We then treated the fragment with the second antibody and were able to start assembling a vocabulary of two-tag combinations.” Since a single DNA molecule was being analyzed each time, the method picked up real-tag interactions even when the source material was as complex as a chunk of brain tissue.
Using mouse embryonic stem cells and tissue samples from the mouse brain and other organs, the scientists applied co-Chip to study the interactions among 14 epigenetic tags, amounting to 70 different combinations. In addition to confirming that the tags indeed occur together on the same DNA molecule, the study identified previously unknown interactions. For example, two interacting tags that the researchers termed “super-enhancers” were found to control the expression of key developmental genes.
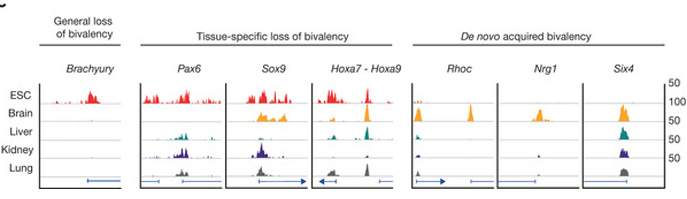
A search for bivalent epigenetic tagging on genes in various tissue types reveals distinct patterns for each. Embryonic stem cells (ESC), in particular, undergo dramatic changes in both amounts and tag pairs as they mature into adult cells.
“Our technology provides proof that the epigenetic code contains complex ‘phrases’ rather than just individual ‘words,’” says Lara-Astiaso. “We can now identify the two-word phrases. The technology can also be further developed in the future to understand more complex combinations of instructions, perhaps even entire ‘sentences.’ The ultimate goal is to decipher the entire epigenetic code that determines how genes function in a cell.”
The research team also included Vladislav Krupalnik, Ohad Gafni and Dr. Jacob Hanna of the Molecular Genetics Department, as well as Eyal David and Dr. Deborah Winter of the Immunology Department.
Dr. Ido Amit’s research is supported by the Benoziyo Endowment Fund for the Advancement of Science; the David and Fela Shapell Family Foundation INCPM Fund for Preclinical Studies; the Wolfson Family Charitable Trust; the Leona M. and Harry B. Helmsley Charitable Trust; and the Rosenwasser Fund for Biomedical Research. Dr. Amit is the incumbent of the Alan and Laraine Fischer Career Development Chair.

Recent Comments