5 in PNAS
24.10.2019
1 – An investigation into the way that double-stranded DNA unwinds for replication, repair or transcription, coauthored by Prof. Yaakov Levy of the Structural Biology Department, together with researchers in Italy, the UK and Spain. By combining simulations and experiments, physics, chemistry and biology, this work reveals that asymmetric base pair dynamics drive the stepwise separation of nucleic acid duplexes. The data suggest a layer of regulation of the genetic material encoded in the “unwindability” of the double helix.
2 – Led by the groups of Dr. Efi Efrati of the Physics of Complex Systems Department and Prof. Ziv Reich of the Biomolecular Sciences Department, this study reveals how the two different domains in the photosynthetic membranes in plants are connected. The intricate structure of the membrane network, which hosts the primary steps of photosynthesis in plant chloroplasts has intrigued scientists for decades. Using electron tomography to determine this structure, the researchers found that the network is consolidated by arrays of right- and left-handed helical structures. Similar arrangements of pitch-balanced helical elements of alternating handedness were proposed to be present in the endoplasmic reticulum and, remarkably, in ultra-dense nuclear matter present in neutron stars and collapsed supernovae. These arrays thus likely represent a universal means to connect between densely packed layers or sheets.
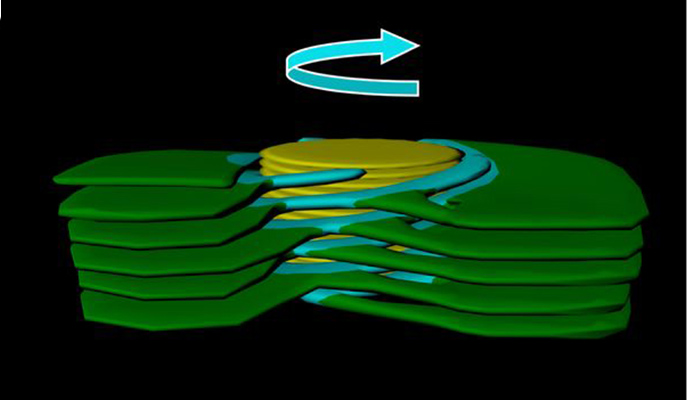
Right-handed helix (from Bussi, et al.)
3 – Prof. Shmuel Pietrokovski of the Molecular Genetics Department identified a novel type of protein introns (inteins) and, together with a group in Germany, showed they can join together with other proteins in conditions in which this has, until now, been impossible. Specifically, these new inteins can modify antibodies and similar proteins without disrupting their native structure. This is likely to have many experimental uses in chemical biology, protein chemistry, and biotechnology.
4 – Research led by the group of Prof. Ada Yonath of the Structural Biology Department, conducted together with researchers in Denmark and Australia, reveals a new possible target for antibiotics in the ribosome of a bacterium, Pseudomonas aeruginosa. Comparing ribosome structures revealed with cryo-electron microscopy from resistant and non-resistant strains, the researchers identified a crucial complex that may be altered to interfere with protein synthesis in the bacterium.
5—Prof. Gad Galili and his group in the Plant and Environmental Sciences Department teamed up with researchers in France to investigate how an RNA suppressor of a virus that attacks plants initiates a molecular pathway that results in autophagy.

Recent Comments