A new study reveals how our microbiomes affect the timing of liver activity
The bacteria we carry around with us in our digestive tract have an influence that spans far beyond the lining of our guts. In research recently published in Cell, a collaborative group at the Weizmann Institute of Science has connected the dots between gut bacteria, their changing behavior over the course of day and night, and the biological clocks dictating the activity in our bodies. In a previous study, this research group had shown that disrupting the host’s biological clock or regular feeding schedules causes changes to the normal daily cyclic behavior of gut bacteria populations. In the present study, the group turned the question around, asking: Do the daily cycles of gut bacterial activity affect that of their hosts, and, if so, how is this accomplished and what is its significance?
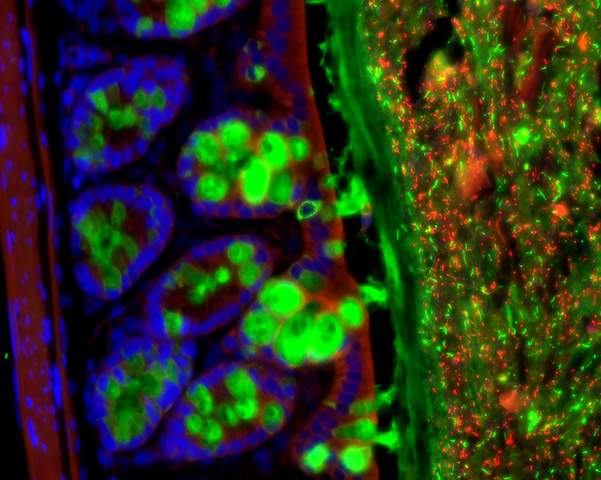
The research, a collaboration between the groups of Dr. Eran Elinav of the Weizmann Institute of Science’s Immunology Department and Prof. Eran Segal of the Applied Mathematics and Computer Science Department, began by disrupting the gut’s microbial population in mice, administering antibiotics that, among other things, disrupted the periodic cycles of microbial activity. The researchers then investigated the effects of this disruption on the daily – or circadian – cycles of gene activity in the host cells lining the intestinal tract. Some of those host genes stuck to their normal circadian cycles, despite the disruption to the bacterial balance. But in another group of genes the cyclic activity was halted; while a third group of genes that had not previously shown periodic behavior now took up cyclic activity.
Research students Christoph Thaiss and Maayan Levi of Elinav’s lab led the research. Collaborating with research student Tal Korem in Segal’s group, the team discovered the mechanism by which the bacteria affect their host’s circadian activity. The changes could be seen hour-by-hour, and they involved alterations to the regulatory activities in the cells that enable the genes to be turned on and off.

The research group next looked at the liver – an organ that has its own diverse sets of tightly coordinated biological clocks. Beginning with either germ-free mice or those treated with antibiotics to disrupt their intestinal bacterial cycles, the group discovered a very similar pattern of liver circadian gene activity to the one they had seen in gut cells: Some liver genes were not affected by disruption of the gut microbiome, but hundreds of other genes lost their daily “rhythm,” or took up a new rhythm following the disruption of the normal gut bacteria function.
How do tiny bacteria affect the timing of activities so far away from home? The answer, the scientists discovered, lies in metabolites – small molecules involved in the exchange of substances throughout the body. Some metabolites are secreted in cycles that are in tune with those of the gut bacteria, and these are passed from the digestive tract to the blood, and from there to such organs as the liver. When the cycles of bacterial activity in the gut were interrupted, certain blood metabolites also exhibited disruptions to their daily cycles. “Even though it has no microbiome of its own, the liver is still affected by the gut bacteria, through their effect on the timing of the secretion of these small molecules,” says Elinav.
What is the medical significance of this finding? To further investigate, the researchers gave the mice paracetamol, a drug that is commonly used by millions and is normally broken down in the liver, but in overdose can cause life-threatening poisoning and liver damage. The researchers noted that in normal mice, the pace at which this drug gets broken down differs when the doses are given at night or during the day. When they disrupted the gut microbiota, the scientists found that that the drug was less likely to be toxic, and its levels were low and remained fixed throughout the day. In other words, interfering in gut bacteria cycles might affect the rate of drug break-down in the liver, and thus possibly, its toxicity.
Even though it has no microbiome of its own, the liver is still affected by the gut bacteria
Segal: “These findings might also suggest the mechanisms for systemic antibiotic side effects that have remained a mystery until now, for example a connection that has been noted between the administration of antibiotics to children and their tendency to obesity as adults.”
Elinav: “We discovered a fascinating ‘tango’ taking place between the gut bacteria and the body’s rhythmic clock, each one leading the other around the daily cycles of life.” “We now know that this coordination between the various clocks means that interfering with the bacterial balance in one place can have far-reaching effects in others,” adds Segal.
Also participating in this research were Dr. Meirav Pevsner-Fischer, Dr. Mally Dori-Bachash and Dr. Hagit Shapiro, and Lenka Dophnalova, Evgeny Tatirovsky, Timur Tuganbaev, Dr. Sara Federici, and Dr. Niv Zmora of Elinav’s lab; David Zeevi of Segal’s lab; Prof. Ido Amit and members of his lab including Dr. Diego Jaitin, Eyal David, Deborah Winter and Meital Gury-Ben Ari; Dr. Elena Kartveshvily of Chemical Research Support; Dr. Alexander Brandis of the Life Sciences Core Facilities; Prof. alon Harmelin of Veterinary Services; Profs. Oren Shibolet and Zamir Halperin of Sourasky Medical Center; and Prof. Kenya Honda of the RIKEN Center Integrative Medical Science, Japan.
Dr. Eran Elinav’s research is supported by the Leona M. and Harry B. Helmsley Charitable Trust; the Adelis Foundation; the Else Kroener Fresenius Foundation; John L. and Vera Schwartz, Pacific Palisades, CA; the Rising Tide Foundation; Andrew and Cynthia Adelson, Canada; Yael and Rami Ungar, Israel; Donald L. Schwarz, Sherman Oaks, CA; Leesa Steinberg, Canada; Valerie and Aaron Edelheit, Santa Barbara, CA; Jack N. Halpern, New York, NY; and the Lawrence and Sandra Post Family. Dr. Elinav is the Incumbent of the Rina Gudinski Career Development Chair.
Prof. Eran Segal’s research is supported by the Crown Human Genome Center, which he heads; the Else Kroener Fresenius Foundation; Donald L. Schwarz, Sherman Oaks, CA; Jack N. Halpern, New York, NY; and Leesa Steinberg, Canada.

Recent Comments