Pairing cells makes the smaller ones easier to study
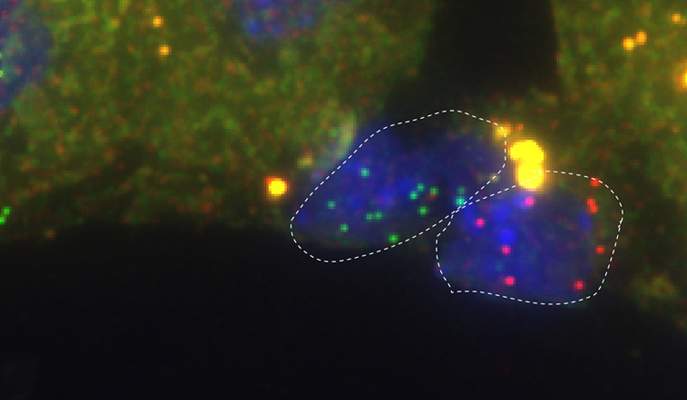
Two endothelial cells in the liver: the cell on the right expresses high levels of the Wnt2 gene (red), and the one on the left, high levels of Dkk3, the “anti-Wnt” gene (green)
“Tell me who your friends are, and I’ll tell you who you are” turns out to be a good guideline for cell research. Researchers at the Weizmann Institute of Science have managed to map out different subtypes of tiny liver cells by pairing them up with their “buddies” – adjacent larger cells that are easier to study. As described in a recent issue of Nature Biotechnology, the pairing-up method may be applied to the mapping of other cell types in the body, and in the future it may facilitate tissue engineering and thedevelopment of new therapies.
Scientists generally map out cell subtypes in various organs by studying their mRNAs, messenger molecules that contain genetic instructions for making proteins. In earlier work, Dr. Shalev Itzkovitz of the Molecular Cell Biology Department had revealed that liver cells called hepatocytes – the liver’s main “work-horses,” which enable this organ to filter out toxins and perform other vital tasks – can be divided into at least nine subtypes, each occupying a different location and specializing in its own tasks.
Such cells are more than mere helpers
But other liver cells – for example, such supporting cell types as endothelial cells – are harder to map. These cells serve as the building blocks of blood vessels and provide an interface between blood vessels and hepatocytes. Such cells are more than mere helpers; they play regulating roles in liver tissue. Since they are so small, however – about 20 times smaller than hepatocytes – individual cells don’t contain enough mRNA to enable researchers to define their locations and characteristics.

(l-r) Adi Egozi, Dr. Keren Bahar Halpern, Dr. Hassan Massalha, and Dr. Shalev Itzkovitz look for liver cells by finding their friends
In the new study, Itzkovitz and his colleagues got around this problem by removing pairs of cells from liver-tissue extract, each made of an endothelial cell attached to a hepatocyte. They then proceeded in two steps. First, relying on their earlier study, they mapped out the different subtypes of hepatocytes in the pairs. Next, they sampled single endothelial cells from different locations and analyzed their genomes, identifying more than a thousand genes that were exclusively expressed in endothelial cells and not in hepatocytes. This enabled them to tell which mRNAs in each cell pair had originated from the endothelial cell and not from its neighbor hepatocyte.
They then collected entire groups of endothelial cells from each tissue location, obtaining enough mRNA to define the genomic profile of each subgroup. They found that expression levels of about a third of the endothelial cell genes increase or decrease gradually as one moves from one subtype of cell to another along a blood vessel wall.
Subtypes spell tight control
The mapping has already produced an important revelation. It had been known that a subtype of endothelial cells enveloping a major vein in the liver has high expression levels of a gene called Wnt, which regulates important genes in adjacent hepatocytes. Surprisingly, the scientists discovered that next to these is another subtype: endothelial cells expressing high levels of another gene that acts as an anti-Wnt, shutting down the Wnt reaction chain. It’s possible that the two cell subtypes emit On and Off signals that together control the activity of hepatocytes.
“Our pairing-up method can be applied to mapping out subtypes of small, supporting cells in the brain, kidneys, heart and various other body organs,” says Itzkovitz. Ultimately, the method can help create an atlas of all human cells, whose number stands at about 100 trillion – 1 followed by 14 zeroes.
Ultimately, the method can help create an atlas of all human cells
Mapping subtypes on this level of detail might provide a way forward for tissue engineering, particularly when it comes to such a highly structured organ as the liver. If, in the future, scientists learn how to engineer artificial portions of the liver, they will need to place the appropriate supporting cells in a way that mimics living liver tissue. Artificial liver tissue may be used for transplantation purposes, reducing the need for donor livers; it could also be used in drug development for testing the toxicity of various drugs – tests that are currently performed in laboratory animals.

Expression levels of a particular gene (green dots) increase gradually (left to right) in endothelial cells lining a blood vessel in the liver
Mapping out endothelial cell subtypes may also advance the development of new therapies for such diseases as fibrosis, in which scar tissue forms in the liver. Understanding the contributions of different cell subtypes to this process may help interfere with the scar formation. Insights gained through the new method may also help develop anticancer drugs aimed at the supporting cells that facilitate tumor growth.
Study authors included: Dr. Keren Bahar Halpern, Rom Shenhav, Dr. Hassan Massalha, Dr. Beata Toth, Adi Egozi, Efi E. Massasa and Dr. Andreas Moor of the Molecular Cell Biology Department; Prof. Ido Amit, Dr. Chiara Medaglia, Eyal David and Amir Giladi of the Immunology Department; and Dr. Ziv Porat of the Life Sciences Core Facilities Department.
Dr. Shalev Itzkovitz’s research is supported by the Wolfson Family Charitable Trust; the Edmond de Rothschild Foundations; and the European Research Council. Dr. Itzkovitz is the incumbent of the Philip Harris and Gerald Ronson Career Development Chair.

Recent Comments